<< back
To perform detailed characterisation of our mouse models and patient samples, we perform metabolomics, lipidomics and proteomics analysies (OMICS analysis) using different mass spectrometry methods.
Recently, we have established mass spectrometry imaging (MSI) and AI/ML-assisted segmentation of MS spectra together with location tensor information to detect new insides in brain diseases. Currently, we perform metabolomics and lipidomics analysis to highlight the function of lipid-transporting ABC transporters, such as ABCA1, ABCA2, ABCA4, and ABCA7. These transporters are risk factors for metabolic and lipid changes in the brain and promote disease development of Alzheimer’s disease and other neurodegenrative disease.
* Projects:
Read more about PROP-AD funded JPND project….
* Project application links:
Read more about ABComics, non-funded application to JPND 2023….
Read more about MSIpattern, non-funded application to JPND 2017….
Read more about A7HD, non-funded application to JPND 2024….
* Norwegian Network of Advanced Proteomics Infrastructure:
Read more about NAPI network….
* Examples of MS imaging and Deep Learning technology for the analysis of mouse (own dataset) and human brain tissue:
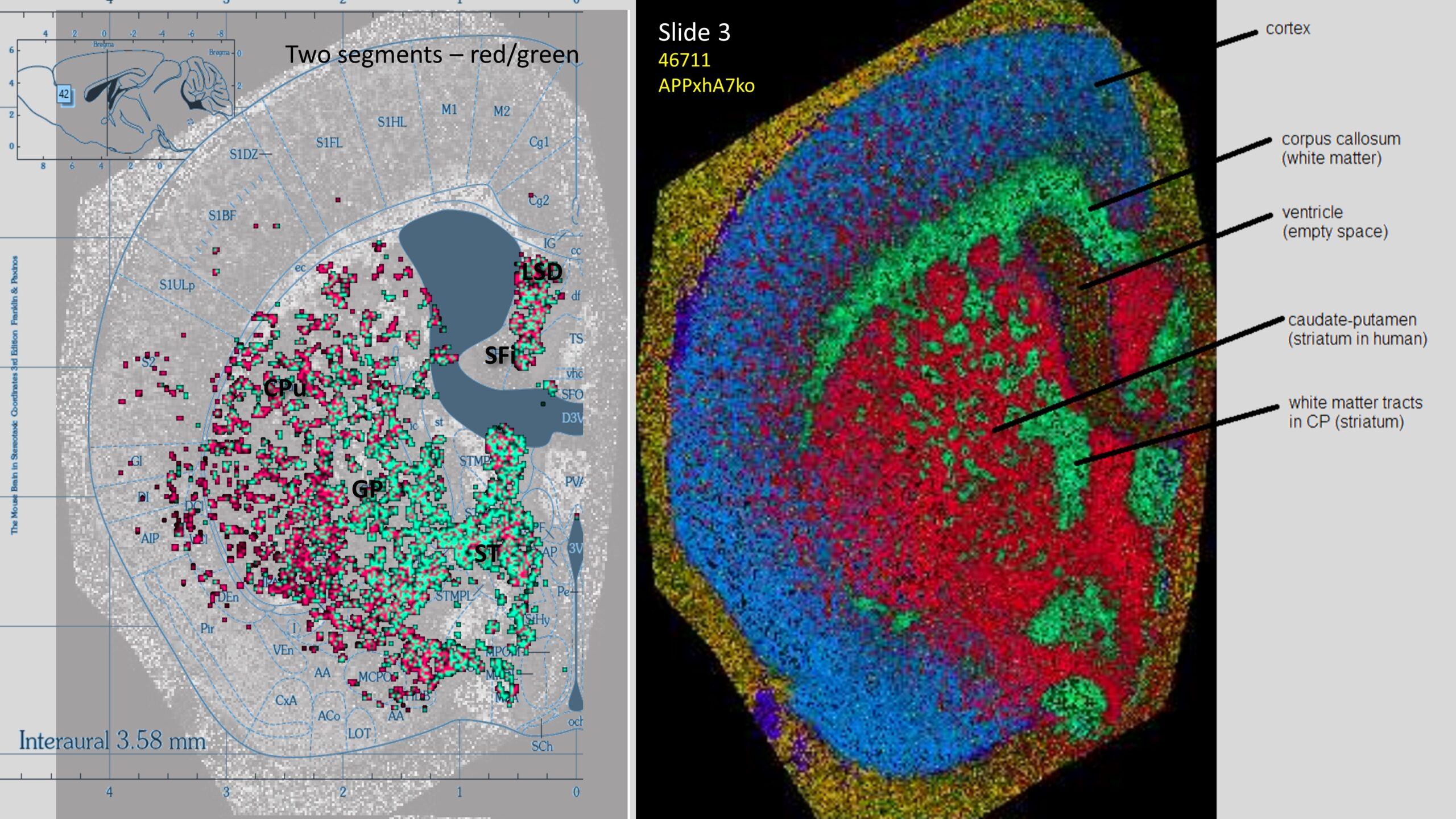 Segmentation of a mouse hemisphere (lipid MSI 300-1350Da, NEDC, 20µm res) showing cortex (blue), white matter (green) and basal ganglia (CPu + GP, red).
Segmentation of a mouse hemisphere (lipid MSI 300-1350Da, NEDC, 20µm res) showing cortex (blue), white matter (green) and basal ganglia (CPu + GP, red).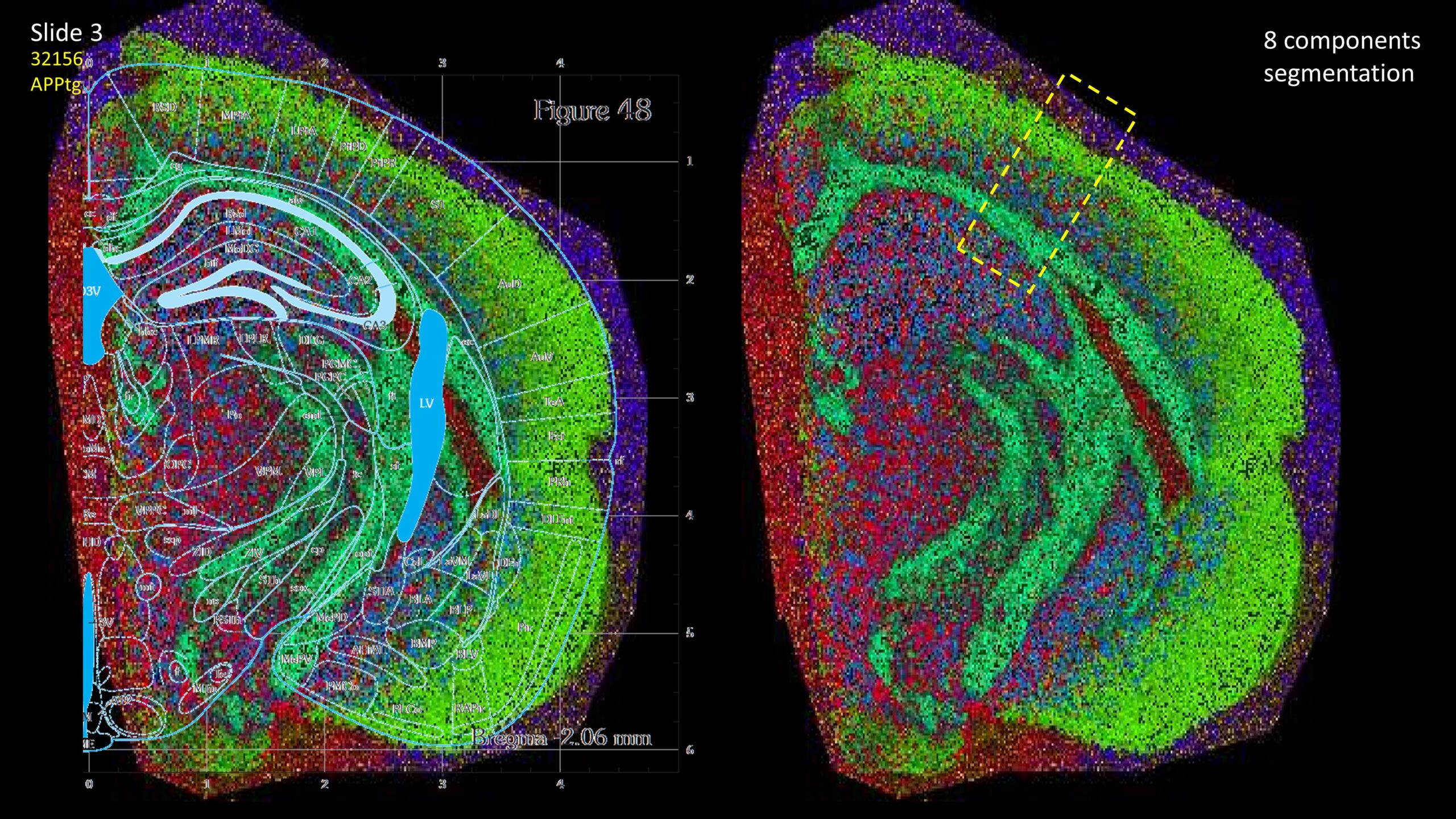
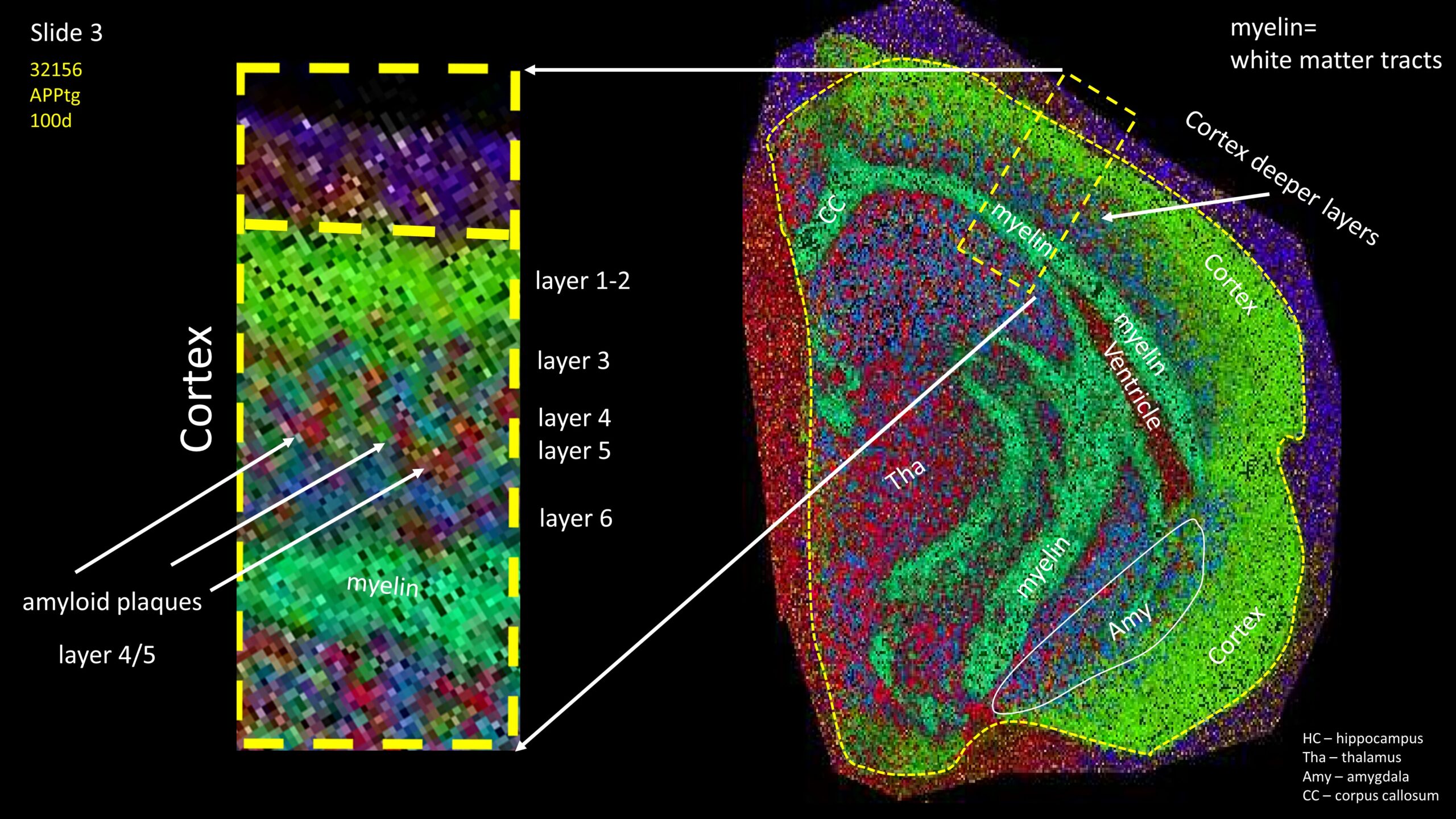 Segmentation of a mouse hemisphere (lipid MSI 300-1350Da, NEDC, 20µm res) showing correct segmentation of cortex and cortical ‘lipids holes’ resembling beta-amyloid plaques in this APPtg model.
Segmentation of a mouse hemisphere (lipid MSI 300-1350Da, NEDC, 20µm res) showing correct segmentation of cortex and cortical ‘lipids holes’ resembling beta-amyloid plaques in this APPtg model.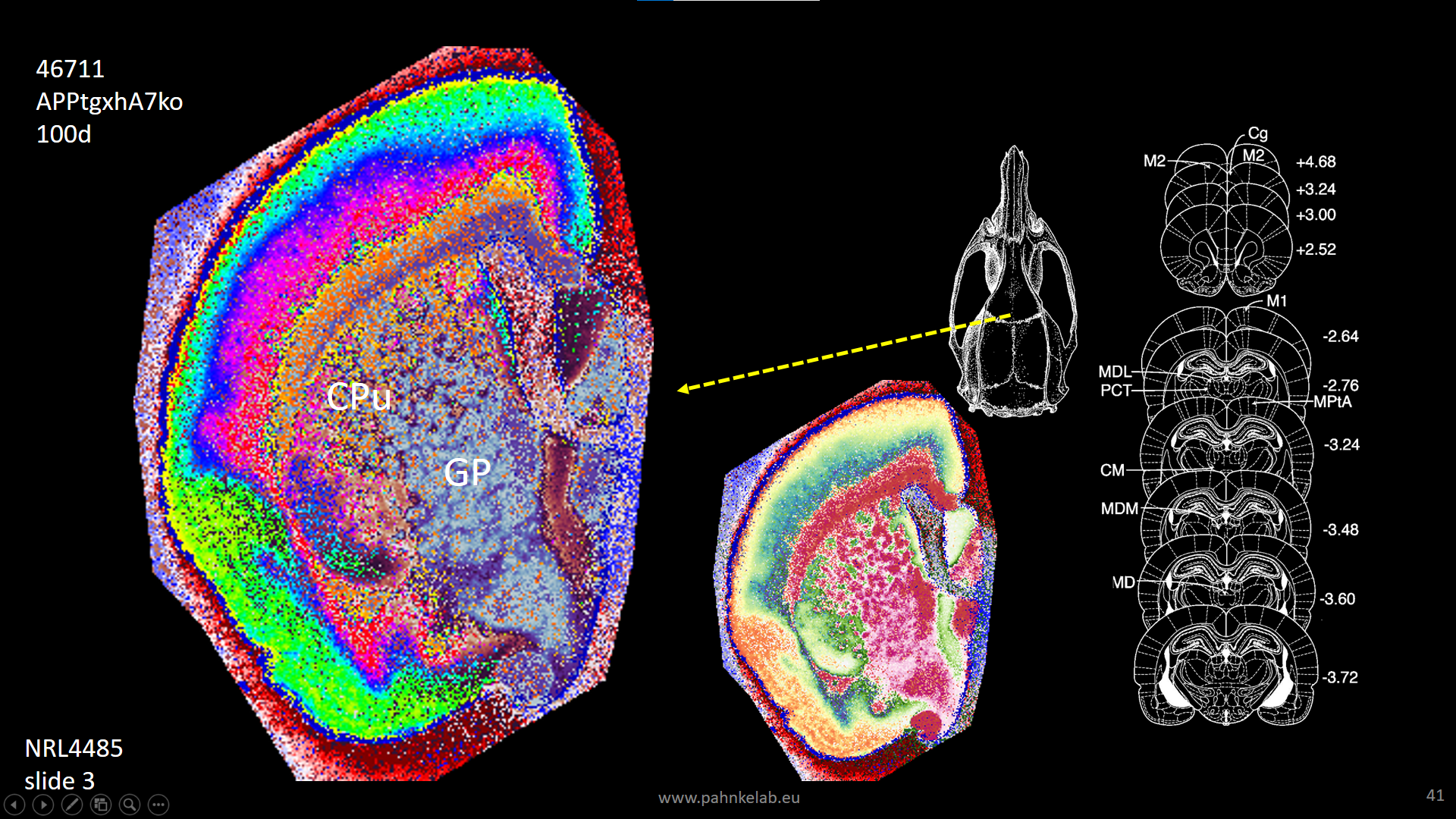
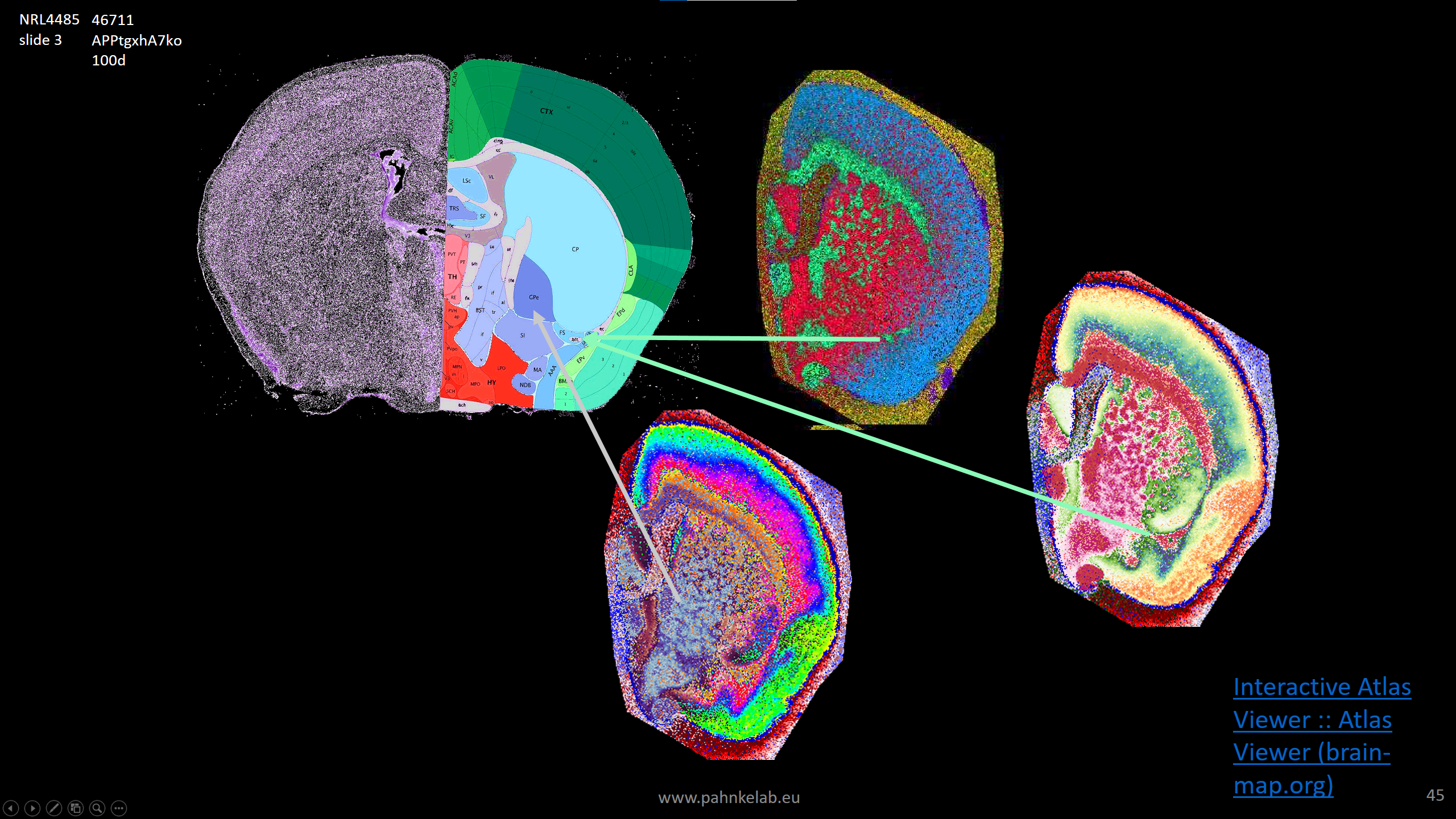
 Segmentation of a mouse hemisphere (lipid MSI 300-1350Da, NEDC, 20µm res) showing correct segmentation of hippocampal structures.
Segmentation of a mouse hemisphere (lipid MSI 300-1350Da, NEDC, 20µm res) showing correct segmentation of hippocampal structures.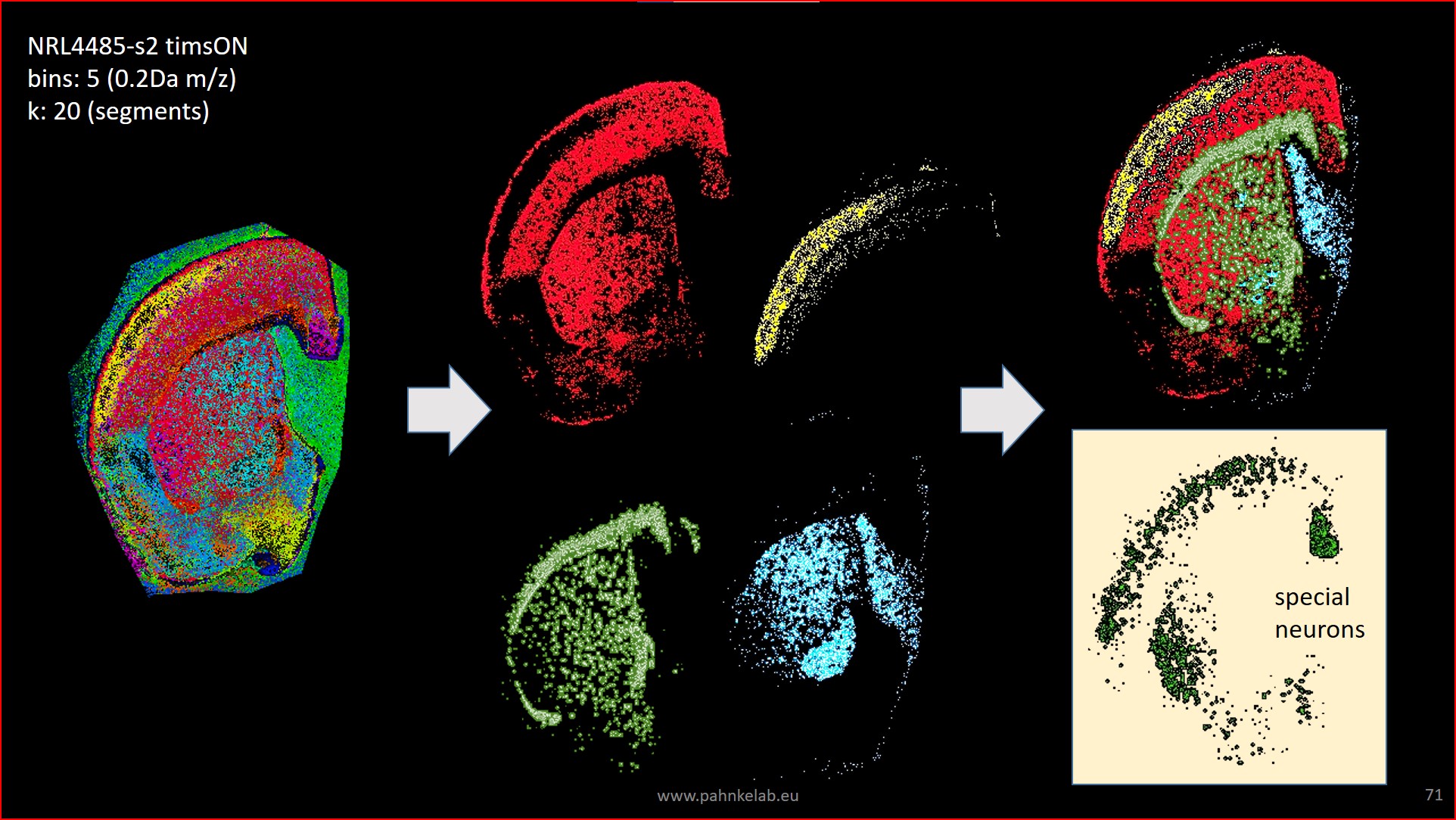
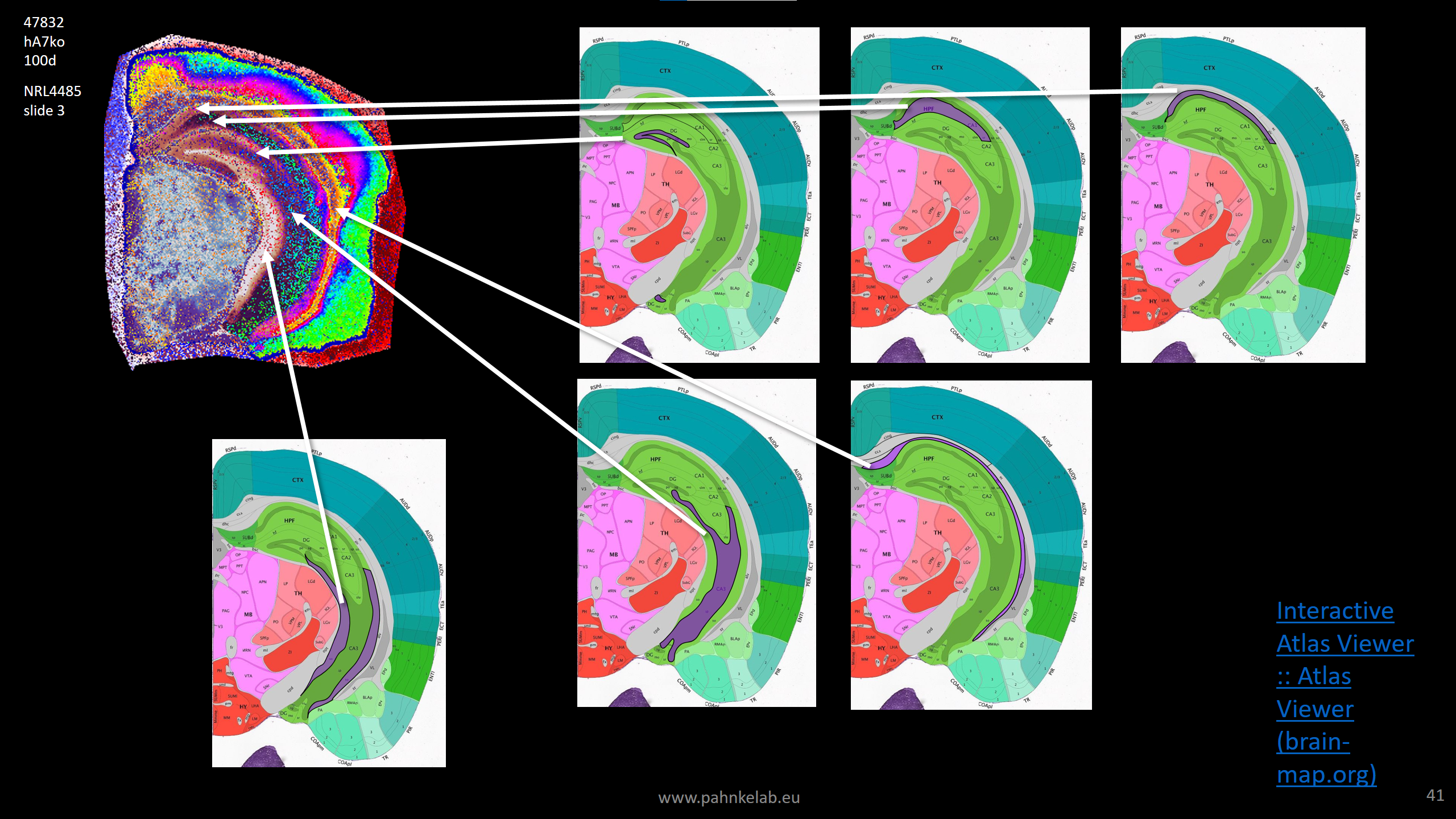
 Segmentation of human cortex based on lipid MSI of a AD patient (sporadic or familiar unknown, UCL brain bank, UK; scan data by Enzlein et al. (2024) AnalChem – Pride dataset PXD049325, lipid MSI 300-2000Da).
Segmentation of human cortex based on lipid MSI of a AD patient (sporadic or familiar unknown, UCL brain bank, UK; scan data by Enzlein et al. (2024) AnalChem – Pride dataset PXD049325, lipid MSI 300-2000Da). Detection of amyloid plaques in a APPtg mouse hemisphere (scan resolution 20µm/pixel).
Detection of amyloid plaques in a APPtg mouse hemisphere (scan resolution 20µm/pixel).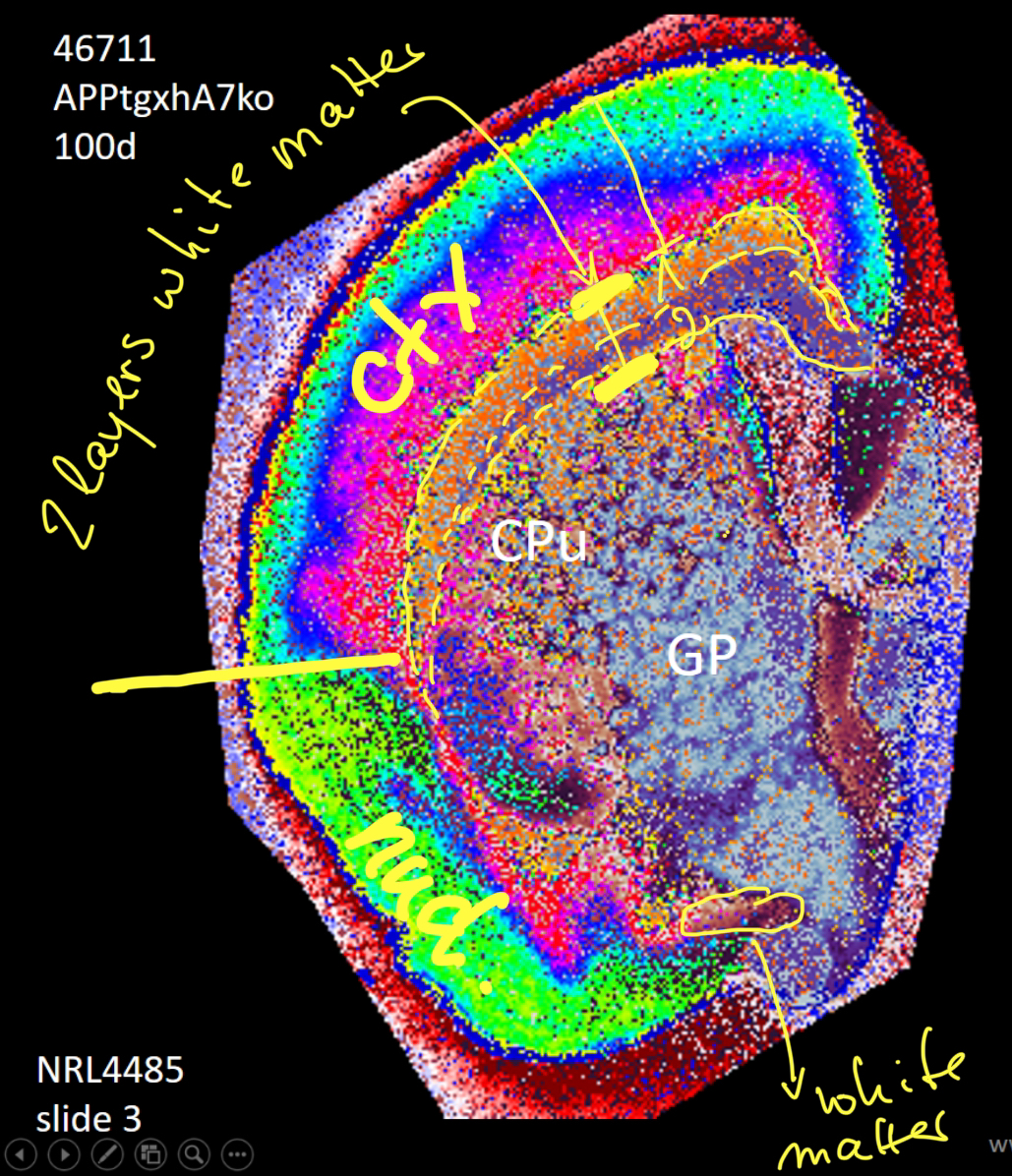
* Published Data Sets:
- Gorska et al. 2024 – J Neurosci
OSF.io: 10.17605/OSF.IO/VWQ58
ProteomeXchange: PXD048864 - Brackhan et al. 2022 – EMBO reports
OSF.io: 10.17605/OSF.IO/VNGPM
MassIVE: 10.25345/C57H1DR1J
Peptide Atlas: PASS01757
* Published Research Papers (selection):
-
Evaluation of cerebrospinal fluid (CSF) and interstitial fluid (ISF) mouse proteomes for the validation and description of Alzheimer’s disease biomarkers
J Neurosci Meth (2024)
Górska AM, Santos-Garcia I, Eiriz I, Brüning, Nyman TA, Pahnke J
PubMed PDF
Open Data Access , PRIDE dataset
-
Isotope-labeled amyloid-β does not transmit to the brain in a prion-like manner after peripheral administration
EMBO reports (2022)
Brackhan M, Calza G, Lundgren K, Bascuñana P, Brüning T, Soliymani R, Kumar R, Abelein A, Baumann M, Lalowski M, and Pahnke J
23(7):e54405 PubMed PDF
-
2-DE profiling of GDNF overexpression-related proteome changes in differentiating ST14A rat progenitor cells.
Proteomics (2007)
Hoffrogge R, Beyer S, Hubner R, Mikkat S, Mix E, Scharf C, Schmitz U, Pauleweit S, Berth M, Zubrzycki I, Christoph H, Pahnke J, Wolkenhauer O, Uhrmacher A, Volker U, Rolfs A
7(1):33-46 PubMed
-
2-DE proteome analysis of a proliferating and differentiating human neuronal stem cell line (ReNcell VM).
Proteomics (2006)
Hoffrogge R, Mikkat S, Scharf C, Beyer S, Christoph H, Pahnke J, Mix E, Berth M, Uhrmacher A, Zubrzycki IZ, Miljan E, Völker U, Rolfs A
6(6):1833-47 PubMed
-
Collection of soluble variants of membrane proteins for transcriptomics and proteomics
In Silico Biol (2005)
Moeller S, Mix E, Blueggel M, Serrano-Fernandez P, Koczan D, Kotsikoris V, Kunz M, Watson M, Pahnke J, Illges H, Kreutzer M, Mikkat S, Thiesen HJ, Glocker MO, Zettl UK, Ibrahim SM
5, 295-311 PubMed